pyxu.util#
Table of Contents
pyxu.util.array_module#
- get_array_module(x, fallback=None)[source]#
Get the array namespace corresponding to a given object.
- Parameters:
x (
object) – Any object compatible with the interface of NumPy arrays.fallback (
ArrayModule) – Fallback module ifxis not a NumPy-like array. Default behaviour: raise error if fallback used.
- Returns:
namespace – The namespace to use to manipulate
x, orfallbackif provided.- Return type:
- redirect(i, **kwargs)[source]#
Change codepath for supplied array backends.
Some functions/methods cannot be written in module-agnostic fashion. The action of this decorator is summarized below:
Analyze an array-valued parameter (
x) of the wrapped function/method (f).If
xlies in one of the supplied array namespaces: re-route execution to the specified function.If
xlies in none of the supplied array namespaces: executef.
- Parameters:
i (
VarName) – name of the array-like variable infto base dispatch on.kwargs (
Mapping) –key[
str]: array backend short-name as defined inNDArrayInfo.value[
collections.abc.Callable]: function/method to dispatch to.
- Return type:
Notes
Auto-dispatch via
redirect()assumes the dispatcher/dispatchee have the same parameterization, i.e.:if
fis a function -> dispatch possible to another callable with identical signature (i.e., function or staticmethod)if
fis a staticmethod -> dispatch possible to another callable with identical signature (i.e., function or staticmethod)if
fis an instance-method -> dispatch to another instance-method of the class with identical signature.
Example
def f(x, y): return "f" @redirect('x', NUMPY=f) # if 'x' is of type NDArrayInfo.NUMPY, i.e. has def g(x, y): return "g" # short-name 'NUMPY' -> reroute execution to `f` x1 = np.arange(5) x2 = da.array(x1) y = 1 g(x1, y), g(x2, y) # 'f', 'g'
- compute(*args, mode='compute', **kwargs)[source]#
Force computation of Dask collections.
- Parameters:
*args (
object,list) – Any number of objects. If it is a dask object, it is evaluated and the result is returned. Non-dask arguments are passed through unchanged. Python collections are traversed to find/evaluate dask objects within. (Usetraverse=False to disable this behavior.)mode (
str) – Dask evaluation strategy: compute or persist.**kwargs (
dict) – Extra keyword parameters forwarded todask.compute()ordask.persist().
- Returns:
*cargs – Evaluated objects. Non-dask arguments are passed through unchanged.
- Return type:
pyxu.util.complex#
- as_real_op(A, dim_rank=None)[source]#
View complex-valued linear operator as its real-valued equivalent.
Useful to transform complex-valued matrix/vector products to their real-valued counterparts.
- Parameters:
- Returns:
A_r – (N1,…,NK,2, M1,…,MD,2) real-valued equivalent.
- Return type:
Examples
import numpy as np import pyxu.util.complex as cpl codim_shape = (1,2,3) dim_shape = (4,5,6,7) dim_rank = len(dim_shape) rng = np.random.default_rng(0) A = rng.standard_normal((*codim_shape, *dim_shape)) \ + 1j * rng.standard_normal((*codim_shape, *dim_shape)) # (1,2,3 |4,5,6,7 ) A_r = cpl.as_real_op(A, dim_rank=dim_rank) # (1,2,3,2|4,5,6,7,2) x = rng.standard_normal(dim_shape) \ + 1j * rng.standard_normal(dim_shape) # (4,5,6,7 ) x_r = cpl.view_as_real(x) # (4,5,6,7,2) y = np.tensordot(A, x, axes=dim_rank) # (1,2,3 ) y_r = np.tensordot(A_r, x_r, axes=dim_rank+1) # (1,2,3,2) np.allclose(y, cpl.view_as_complex(y_r)) # True
Notes
Real-valued matrices are returned unchanged.
See also
- require_viewable(x)[source]#
Copy array if required to do real/complex view manipulations.
Real/complex view manipulations are feasible if the last axis is contiguous.
- view_as_complex(x)[source]#
View real-valued array as its complex-valued bijection. (Inverse of
view_as_real().)- Parameters:
x (
NDArray) – (…, N, 2) real-valued array.- Returns:
y – (…, N) complex-valued array.
- Return type:
Examples
from pyxu.util import view_as_real, view_as_complex x = np.array([[0., 1], [2 , 3], [4 , 5]]) y = view_as_complex(x) # array([0.+1.j, 2.+3.j, 4.+5.j]) view_as_real(y) == x # True
Notes
Complex-valued inputs are returned unchanged.
See also
- view_as_real(x)[source]#
View complex-valued array as its real-valued bijection. (Inverse of
view_as_complex().)- Parameters:
x (
NDArray) – (…, N) complex-valued array.- Returns:
y – (…, N, 2) real-valued array.
- Return type:
Examples
from pyxu.util import view_as_real, view_as_complex x = np.r_[0+1j, 2+3j, 4+5j] y = view_as_real(x) # array([[0., 1.], # [2., 3.], # [4., 5.]]) view_as_complex(y) == x # True
Notes
Real-valued inputs are returned unchanged.
See also
pyxu.util.misc#
- copy_if_unsafe(x)[source]#
Copy array if it is unsafe to do in-place updates on it.
In-place updates are unsafe if:
the array is read-only, OR
the array is a view onto another array.
- import_module(name, fail_on_error=True)[source]#
Load a Python module dynamically.
- Parameters:
- Return type:
- parse_params(func, *args, **kwargs)[source]#
Get function parameterization.
- Returns:
params – (key, value) params as seen in body of
funcwhen called viafunc(*args, **kwargs).- Return type:
- peaks(x, y)[source]#
Matlab 2D peaks function.
Peaks is a function of two variables, obtained by translating and scaling Gaussian distributions. (See Matlab’s peaks function.)
This function is useful for testing purposes.
- Parameters:
- Returns:
z – Values of the 2D function
peaksat the points specified by the entries ofxandy.- Return type:
Examples
import numpy as np import matplotlib.pyplot as plt from pyxu.util.misc import peaks x = np.linspace(-3, 3, 1000) xx, yy = np.meshgrid(x, x) z = peaks(xx, yy) plt.figure() plt.imshow(z)
(
Source code,png,hires.png,pdf)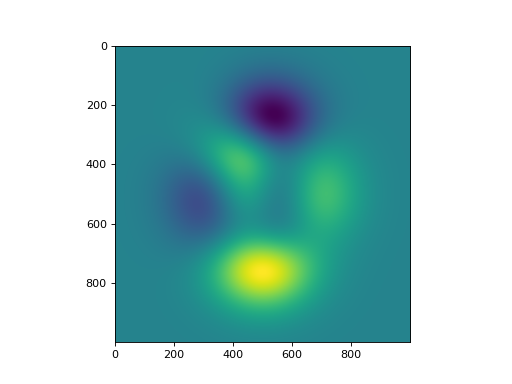
- star_like_sample(N, w, s, po, x0, ndi=NDArrayInfo.NUMPY)[source]#
Star-like test image.
Generates a (N, N) square image of a star-like object normalized between 0 and 1. Based on GlobalBioIm’s StarLikeSample function. This function is useful for testing purposes as it contains high-frequency information.
- Parameters:
N (
Integer) – Size of the image (must be an even number).w (
Integer) – The number of branches of the sample will be 4*w.s (
Real) – Slope of the sigmoid function \(\frac1{1+\exp[s (x-x_{0})]}\) attenuating the boundaries.po (
Integer) – Power-raising factor for the final image (to have smoother edges).x0 (
Real) – Radial shift of the sigmoid function \(\frac1{1+\exp[s (x-x_{0})]}\).ndi (
NDArrayInfo) – Desired array module for the output.
- Returns:
image – (N, N) image of star-like sample.
- Return type:
Examples
import numpy as np import matplotlib.pyplot as plt from pyxu.util.misc import star_like_sample star = star_like_sample(N=256, w=8, s=20, po=3, x0=0.7) plt.figure() plt.imshow(star)
(
Source code,png,hires.png,pdf)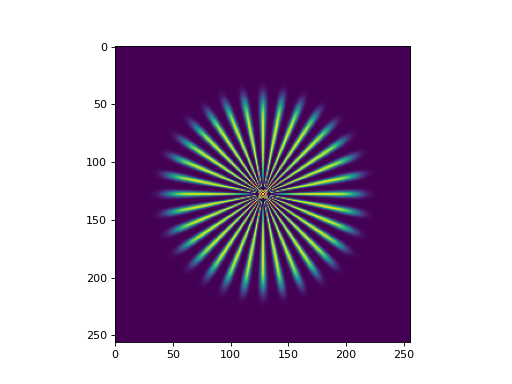
pyxu.util.operator#
- as_canonical_axes(axes, rank)[source]#
Transform NDarray axes into tuple-form with positive indices.
- vectorize(i, dim_shape, codim_shape)[source]#
Decorator to auto-vectorize a function \(\mathbf{f}: \mathbb{R}^{M_{1} \times\cdots\times M_{D}} \to \mathbb{R}^{N_{1} \times\cdots\times N_{K}}\) to accept stacking dimensions.
- Parameters:
i (
VarName) – Function/method parameter to vectorize. This variable must hold an object with a NumPy API.dim_shape (
NDArrayShape) – (M1,…,MD) shape of operator’s domain.codim_shape (
NDArrayShape) – (N1,…,NK) shape of operator’s co-domain.
- Returns:
g – Function/Method with signature
(..., M1,...,MD) -> (..., N1,...,NK)in parameteri.- Return type:
Example
import pyxu.util as pxu def f(x): return x.sum(keepdims=True) N = 5 g = pxu.vectorize("x", N, 1)(f) x = np.arange(2*N).reshape((2, N)) g(x[0]), g(x[1]) # [10], [35] g(x) # [[10], # [35]]
Notes
vectorize()assumes the function being vectorized is thread-safe and can be evaluated in parallel. Using it on thread-unsafe code may lead to incorrect outputs.As predicted by Pyxu’s
OperatorAPI:The dtype of the vectorized function is assumed to match
x.dtype.The array backend of the vectorized function is assumed to match that of
x.